ROS 2
- Open a live connection
- Play back recorded data
Visualize your robot live
The best way to visualize your live ROS 2 robot in Foxglove is using the Foxglove Bridge. It's written using the Foxglove SDK in C++ and is designed for high performance with low overhead. We provide reasonable defaults to help get you up and running quickly, but it's also highly configurable.
We provide multiple ways to install Foxglove Bridge. The steps below assume you want to install from the official ROS package channels using the apt package manager.
1. Install the Foxglove bridge
Install and launch the Foxglove Bridge node:
sudo apt install ros-$ROS_DISTRO-foxglove-bridge
ros2 launch foxglove_bridge foxglove_bridge_launch.xml
The Foxglove Bridge will automatically subscribe to all topics in your ROS system and make them available for live visualization.
For ROS 2 distributions older than Humble, you will have to build from source.
2. Connect to your robot:
Ensure Foxglove Bridge is running.
Make sure you are on the same network as your robot. In Foxglove, select Open connection from the dashboard or left-hand menu.
Select Foxglove WebSocket in the Open a new connection dialog, then enter the URL of the server:
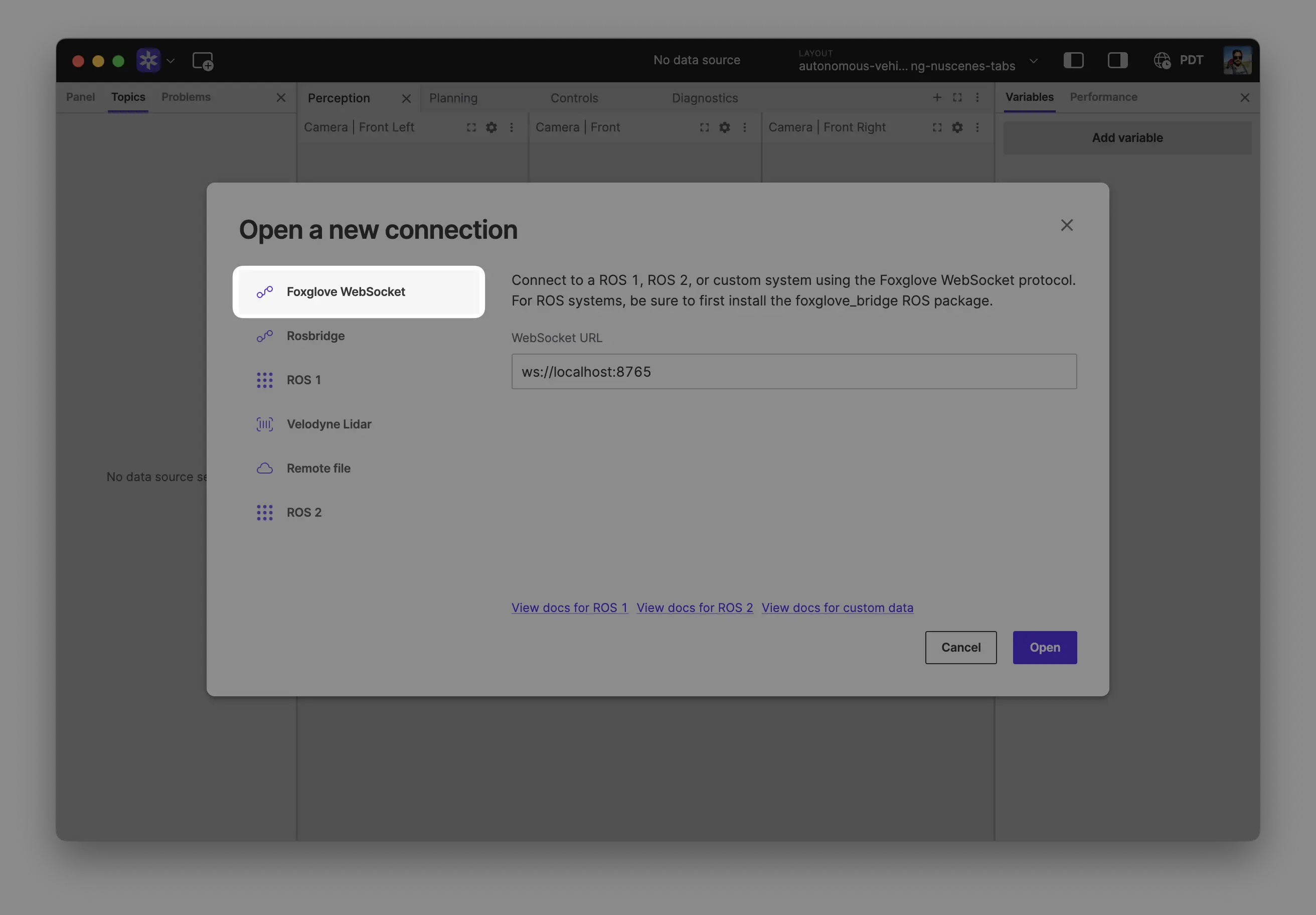
Click "Open" to connect.
Local development: Use ws://localhost:8765 when running the server on the same machine as Foxglove.
Robot connection: Use ws://ROBOT_IP:8765 when connecting to a server running on your robot, where ROBOT_IP is your robot's IP address on the network.
While Foxglove does support rosbridge, we recommend always using the Foxglove Bridge for the best features, performance, and stability.
Opening a MCAP
Foxglove can open MCAP files natively. If you don't have an MCAP file, start by recording data.
1. Record data
With your ROS 2 stack running, use rosbag2 installed as part of your ROS 2 setup to record data:
ros2 bag record -a
2. Open your recording
Open your recording(s) directly from your computer by:
- Dragging and dropping them into Foxglove
- Using
Cmd/Ctrl + o - Clicking Open local file(s) in the app
When opening multiple files, Foxglove will display the data as a single merged timeline. The files must be of the same format.
Older ROS 2 distributions
ROS 2 distributions before Iron produce .db3 files by default. Foxglove can open them, but .db3 does not capture message definitions, meaning custom message types can't easily be played back. MCAP solves this and can be installed on older ROS 2 distributions
sudo apt-get install ros-$ROS_DISTRO-rosbag2-storage-mcap
ros2 bag record -s mcap --all
If you already have .db3 files you want to convert to MCAP use the mcap CLI tool:
mcap convert ros2_input.db3 ros2_output.mcap
mcap will search the path stored in your $AMENT_PREFIX_PATH environment variable to locate the ROS message definitions on your hard drive. You can also specify a colon-separated list of directories for the CLI tool to search using the ament-prefix-path flag:
mcap convert ros2_input.db3 ros2_output.mcap --ament-prefix-path=/your/first/directory;/your/second/directory
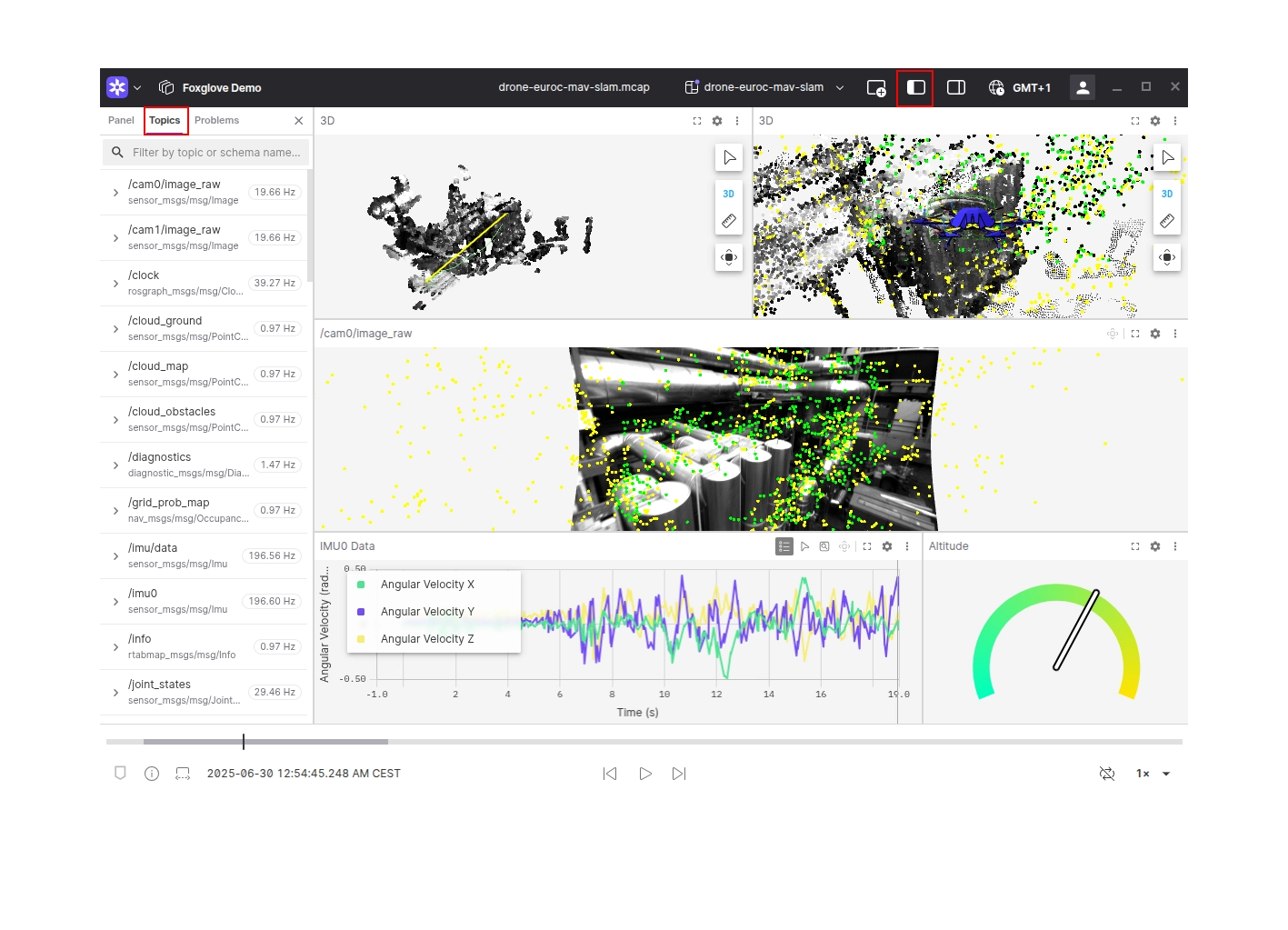
Explore your data
Your data is now available for exploring in Foxglove. Add some panels to begin visualizing what your robot is seeing and producing. Not sure where to start? Try adding a Raw Messages panel, Plot panel, or 3D panel.
To list available topics, open the left sidebar and open the Topics tab.
Learn more
- Explore all of the panels Foxglove has to offer for viewing and exploring data
- Create and save sets of panels and their setting as layouts to reuse them or share with teammates
- Use the data management platform to store, stream, and share your robotics data
- Learn about all the configuration options available for Foxglove Bridge